class: center, middle, inverse, title-slide # Lecture 3 - Data manipulation ## <code>tidyverse</code> intro. 2 ### Issac Lee ### 2021-03-11 --- class: center, middle # Sungkyunkwan University  ## Actuarial Science --- class: center, middle # Part 2: Dive into tidyverse --- class: center, middle # tibble --- # tibble - morden reinterpretation of the `data.frame` .pull-left[ ```r as_tibble(mtcars) ``` ``` ## # A tibble: 6 x 11 ## mpg cyl disp hp drat wt qsec vs ## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 21 6 160 110 3.9 2.62 16.5 0 ## 2 21 6 160 110 3.9 2.88 17.0 0 ## 3 22.8 4 108 93 3.85 2.32 18.6 1 ## 4 21.4 6 258 110 3.08 3.22 19.4 1 ## 5 18.7 8 360 175 3.15 3.44 17.0 0 ## 6 18.1 6 225 105 2.76 3.46 20.2 1 ## # ... with 3 more variables: am <dbl>, ## # gear <dbl>, carb <dbl> ``` ] .pull-right[ ```r tibble(x = 1:2, y = 1, z = x^2 + y) ``` ``` ## # A tibble: 2 x 3 ## x y z ## <int> <dbl> <dbl> ## 1 1 1 2 ## 2 2 1 5 ``` ```r tribble( ~x, ~y, ~z, "a", 2, 3.6 ) ``` ``` ## # A tibble: 1 x 3 ## x y z ## <chr> <dbl> <dbl> ## 1 a 2 3.6 ``` ] --- # tibble vs. dataframe * Looks similar but they are different * Two main differences: printing, and subsetting. .pull-left[ ```r mtcars <- mtcars %>% head() class(mtcars) ``` ``` ## [1] "data.frame" ``` ```r mtcars ``` ``` ## mpg cyl disp hp drat wt ## Mazda RX4 21.0 6 160 110 3.90 2.620 ## Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 ## Datsun 710 22.8 4 108 93 3.85 2.320 ## Hornet 4 Drive 21.4 6 258 110 3.08 3.215 ## Hornet Sportabout 18.7 8 360 175 3.15 3.440 ## Valiant 18.1 6 225 105 2.76 3.460 ## qsec vs am gear carb ## Mazda RX4 16.46 0 1 4 4 ## Mazda RX4 Wag 17.02 0 1 4 4 ## Datsun 710 18.61 1 1 4 1 ## Hornet 4 Drive 19.44 1 0 3 1 ## Hornet Sportabout 17.02 0 0 3 2 ## Valiant 20.22 1 0 3 1 ``` ] .pull-right[ ```r as_tibble(mtcars) ``` ``` ## # A tibble: 6 x 11 ## mpg cyl disp hp drat wt qsec vs ## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 21 6 160 110 3.9 2.62 16.5 0 ## 2 21 6 160 110 3.9 2.88 17.0 0 ## 3 22.8 4 108 93 3.85 2.32 18.6 1 ## 4 21.4 6 258 110 3.08 3.22 19.4 1 ## 5 18.7 8 360 175 3.15 3.44 17.0 0 ## 6 18.1 6 225 105 2.76 3.46 20.2 1 ## # ... with 3 more variables: am <dbl>, ## # gear <dbl>, carb <dbl> ``` ] --- # tibble - strict about subsetting * You cannot access to a variable that doesn't exist. .pull-left[ ```r mtcars %>% names() ``` ``` ## [1] "mpg" "cyl" "disp" "hp" "drat" "wt" ## [7] "qsec" "vs" "am" "gear" "carb" ``` ```r my_df <- mtcars[,1:5] my_df$dis # ?? we don't have dis variable.. ``` ``` ## [1] 160 160 108 258 360 225 ``` ] .pull-right[ ```r my_tb <- mtcars[,1:5] %>% as_tibble() my_tb$dis # yes! we don't have dis variable! ``` ``` ## Warning: Unknown or uninitialised column: `dis`. ``` ``` ## NULL ``` ] --- # tibble - more precise about return values * Sometimes `R` is too much kind. .pull-left[ * convert dataframe into vector automatically ```r class(my_df[,1]) ``` ``` ## [1] "numeric" ``` ```r my_df[,1] ``` ``` ## [1] 21.0 21.0 22.8 21.4 18.7 18.1 ``` ] .pull-right[ * That's nope! in `tibble` ```r class(my_tb[,1]) ``` ``` ## [1] "tbl_df" "tbl" "data.frame" ``` ```r my_tb[,1] ``` ``` ## # A tibble: 6 x 1 ## mpg ## <dbl> ## 1 21 ## 2 21 ## 3 22.8 ## 4 21.4 ## 5 18.7 ## 6 18.1 ``` ] --- # tibble - return value * `[` returns `tibble`, `[[` returns `vector` always .pull-left[ ```r my_tb[,1] ``` ``` ## # A tibble: 6 x 1 ## mpg ## <dbl> ## 1 21 ## 2 21 ## 3 22.8 ## 4 21.4 ## 5 18.7 ## 6 18.1 ``` ] .pull-right[ ```r my_tb[[1]] ``` ``` ## [1] 21.0 21.0 22.8 21.4 18.7 18.1 ``` ] --- # list dataframe * Do you know list can be part of the data.frame? * This will be useful, when you work with `purrr`. .pull-left[ ```r my_df$cyl <- list(9, 10:11, 12:14, "text", as.factor("a"), as.factor("b")) my_df ``` ``` ## mpg cyl disp hp drat ## Mazda RX4 21.0 9 160 110 3.90 ## Mazda RX4 Wag 21.0 10, 11 160 110 3.90 ## Datsun 710 22.8 12, 13, 14 108 93 3.85 ## Hornet 4 Drive 21.4 text 258 110 3.08 ## Hornet Sportabout 18.7 1 360 175 3.15 ## Valiant 18.1 1 225 105 2.76 ``` ] .pull-right[ ```r my_tb$cyl <- list(9, 10:11, 12:14, "text", as.factor("a"), as.factor("b")) my_tb ``` ``` ## # A tibble: 6 x 5 ## mpg cyl disp hp drat ## <dbl> <list> <dbl> <dbl> <dbl> ## 1 21 <dbl [1]> 160 110 3.9 ## 2 21 <int [2]> 160 110 3.9 ## 3 22.8 <int [3]> 108 93 3.85 ## 4 21.4 <chr [1]> 258 110 3.08 ## 5 18.7 <fct [1]> 360 175 3.15 ## 6 18.1 <fct [1]> 225 105 2.76 ``` ] --- class: middle, center # tidyr --- # Example * tibble which is not a `tidydata` ```r tibble_wide <- tribble( ~name, ~gender, ~party, ~"2019", ~"2020", ~"2021", "Bomi Kim", "female", "opposition", 48.01, 56.99, 57.43, "Issac Lee", "male", "ruling", 47.85, 30.92, 42.20, "Soony Kim", "female", "ruling", 33.49, 41.86, 44.54, "Jelly Lee", "female", "opposition", 42.35, 32.20, 40.19 ) ``` --- # `pivot_longer()` intuition  --- # Graph with year vs approval rating ```r tibble_wide %>% pivot_longer( cols = "2019":"2021", names_to = "year", values_to = "approval_rating" ) ``` ``` ## # A tibble: 12 x 5 ## name gender party year approval_rating ## <chr> <chr> <chr> <chr> <dbl> ## 1 Bomi Kim female opposit~ 2019 48.0 ## 2 Bomi Kim female opposit~ 2020 57.0 ## 3 Bomi Kim female opposit~ 2021 57.4 ## 4 Issac Lee male ruling 2019 47.8 ## 5 Issac Lee male ruling 2020 30.9 ## 6 Issac Lee male ruling 2021 42.2 ## 7 Soony Kim female ruling 2019 33.5 ## 8 Soony Kim female ruling 2020 41.9 ## 9 Soony Kim female ruling 2021 44.5 ## 10 Jelly Lee female opposit~ 2019 42.4 ## 11 Jelly Lee female opposit~ 2020 32.2 ## 12 Jelly Lee female opposit~ 2021 40.2 ``` --- # `pivot_longer()` and ggplot comb. .pull-left[ ```r tibble_wide %>% pivot_longer( cols = "2019":"2021", names_to = "year", values_to = "approval_rating" ) %>% ggplot(aes(x = year, y = approval_rating, group = name, color = name)) + geom_path() ``` ] .pull-right[ 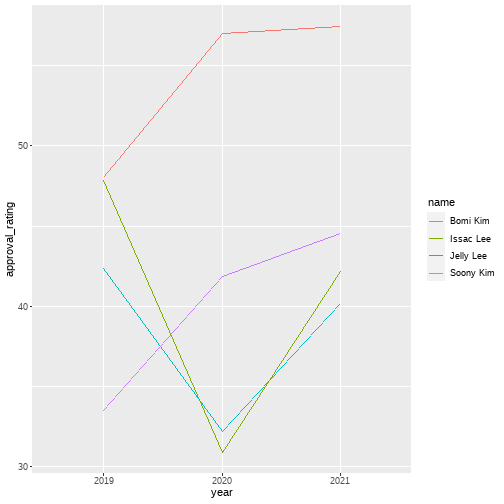<!-- --> ] --- # `pivot_wider()` ```r tibble_long <- tibble_wide %>% pivot_longer( cols = "2019":"2021", names_to = "year", values_to = "approval_rating" ) tibble_long %>% pivot_wider(names_from = year, values_from = approval_rating) ``` ``` ## # A tibble: 4 x 6 ## name gender party `2019` `2020` `2021` ## <chr> <chr> <chr> <dbl> <dbl> <dbl> ## 1 Bomi Kim female opposition 48.0 57.0 57.4 ## 2 Issac Lee male ruling 47.8 30.9 42.2 ## 3 Soony Kim female ruling 33.5 41.9 44.5 ## 4 Jelly Lee female opposition 42.4 32.2 40.2 ``` --- # `rowwise()` and `c_across()` combo. ```r tibble_wide <- tibble_wide %>% rowwise() %>% mutate(average = mean(c_across("2019":"2021"))) tibble_wide ``` ``` ## # A tibble: 4 x 7 ## # Rowwise: ## name gender party `2019` `2020` `2021` average ## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> ## 1 Bomi ~ female oppo~ 48.0 57.0 57.4 54.1 ## 2 Issac~ male ruli~ 47.8 30.9 42.2 40.3 ## 3 Soony~ female ruli~ 33.5 41.9 44.5 40.0 ## 4 Jelly~ female oppo~ 42.4 32.2 40.2 38.2 ``` --- class: center, middle # purrr --- # Two important iteration paradigms * Imperative programming: Loop, while loops.. - Good side: Great place to start, Explicit and obvious - Bad side: Verbose, duplication * Functional programming - Compact, fewer errors - Not for the beginners. :) --- # Functional * Function of functions .pull-left[ ```r odd_sum <- function(vec){ sum(c(vec)[as.logical(vec %% 2)]) } odd_sum(1:100) ``` ``` ## [1] 2500 ``` ```r odd_mean <- function(vec){ mean(c(vec)[as.logical(vec %% 2)]) } odd_mean(1:100) ``` ``` ## [1] 50 ``` ] .pull-right[ ```r odd_functional <- function(vec, fun){ fun(c(vec)[as.logical(vec %% 2)]) } odd_functional(1:100, sum) ``` ``` ## [1] 2500 ``` ```r odd_functional(1:100, mean) ``` ``` ## [1] 50 ``` ] --- # `purrr` is FP toolkit in `tidyverse` * `base` package has `apply`, `lapply`, ... * `purrr` is more consistent, once you understand you can expect its behavior better. - `map()` results a list. - `map_lgl()` results a logical vector. - `map_int()` results an integer vector. - `map_dbl()` results a double vector. - `map_chr()` results a character vector. --- # `map()` basic .pull-left[ ```r df <- tibble( a = rnorm(4), b = rnorm(4), c = rnorm(4), d = rnorm(4) ) df ``` ``` ## # A tibble: 4 x 4 ## a b c d ## <dbl> <dbl> <dbl> <dbl> ## 1 -0.493 0.194 -0.433 1.83 ## 2 1.58 -0.259 -0.420 -0.217 ## 3 0.118 0.526 0.547 -1.29 ## 4 -0.328 -0.138 0.881 1.59 ``` ] .pull-right[ * List result ```r map(df, mean) ``` ``` ## $a ## [1] 0.2184034 ## ## $b ## [1] 0.08073396 ## ## $c ## [1] 0.143805 ## ## $d ## [1] 0.4781553 ``` ] --- # `map_<result>` * `result` should match with expectation * This behavior is actually good for us! .pull-left[ ```r map_dbl(df, mean) ``` ``` ## a b c d ## 0.21840342 0.08073396 0.14380497 0.47815533 ``` ```r map_df(df, mean) ``` ``` ## # A tibble: 1 x 4 ## a b c d ## <dbl> <dbl> <dbl> <dbl> ## 1 0.218 0.0807 0.144 0.478 ``` ] .pull-right[ ```r map_int(df, mean) ``` ``` ## Error: Can't coerce element 1 from a double to a integer ``` ```r map_chr(df, mean) ``` ``` ## a b c d ## "0.218403" "0.080734" "0.143805" "0.478155" ``` ] --- # the `purrr` verse We have two things to remember; * `~`: function * `.`: current list element .pull-left[ ```r map_dbl(df, function(df){ sqrt(mean(df)^2 + 3)}) ``` ``` ## a b c d ## 1.745766 1.733931 1.738010 1.796840 ``` ] .pull-right[ ```r map_dbl(df, ~sqrt(mean(.)^2 + 3)) ``` ``` ## a b c d ## 1.745766 1.733931 1.738010 1.796840 ``` ] --- # `split()` and `map()` comb. ```r library(palmerpenguins) penguins %>% split(.$species) %>% map(~lm(bill_length_mm ~ bill_depth_mm, data = .)) ``` ``` ## $Adelie ## ## Call: ## lm(formula = bill_length_mm ~ bill_depth_mm, data = .) ## ## Coefficients: ## (Intercept) bill_depth_mm ## 23.068 0.857 ## ## ## $Chinstrap ## ## Call: ## lm(formula = bill_length_mm ~ bill_depth_mm, data = .) ## ## Coefficients: ## (Intercept) bill_depth_mm ## 13.428 1.922 ## ## ## $Gentoo ## ## Call: ## lm(formula = bill_length_mm ~ bill_depth_mm, data = .) ## ## Coefficients: ## (Intercept) bill_depth_mm ## 17.230 2.021 ``` --- # Nested data - important! * dataframes in `dataframe`! * Why is this important? .pull-left[ ```r nested_df <- penguins %>% group_by(species) %>% nest() %>% ungroup(species) nested_df ``` ``` ## # A tibble: 3 x 2 ## species data ## <fct> <list> ## 1 Adelie <tibble [152 x 7]> ## 2 Gentoo <tibble [124 x 7]> ## 3 Chinstrap <tibble [68 x 7]> ``` ] .pull-right[ ```r nested_df[[2]][[1]] %>% head() ``` ``` ## # A tibble: 6 x 7 ## island bill_length_mm bill_depth_mm ## <fct> <dbl> <dbl> ## 1 Torgersen 39.1 18.7 ## 2 Torgersen 39.5 17.4 ## 3 Torgersen 40.3 18 ## 4 Torgersen NA NA ## 5 Torgersen 36.7 19.3 ## 6 Torgersen 39.3 20.6 ## # ... with 4 more variables: ## # flipper_length_mm <int>, body_mass_g <int>, ## # sex <fct>, year <int> ``` ] --- # let's `purrr` functions to this nested data ```r nested_df %>% mutate(model = map(data, ~lm(bill_length_mm~bill_depth_mm, data = .))) ``` ``` ## # A tibble: 3 x 3 ## species data model ## <fct> <list> <list> ## 1 Adelie <tibble [152 x 7]> <lm> ## 2 Gentoo <tibble [124 x 7]> <lm> ## 3 Chinstrap <tibble [68 x 7]> <lm> ``` --- # let's `purrr` functions to this nested data ```r nested_df %>% mutate(model = map(data, ~lm(bill_length_mm~bill_depth_mm, data = .))) %>% transmute(species, map_df(model, ~summary(.)$coefficients[,1])) ``` ``` ## # A tibble: 3 x 3 ## species `(Intercept)` bill_depth_mm ## <fct> <dbl> <dbl> ## 1 Adelie 23.1 0.857 ## 2 Gentoo 17.2 2.02 ## 3 Chinstrap 13.4 1.92 ``` --- # map2() - Example 1 * Loops using two arguments .pull-left[ ```r mu_vec <- 1:5 * 3 sigma_vec <- 1:5 * 2 n <- sample(1:10, 5) mu_vec ``` ``` ## [1] 3 6 9 12 15 ``` ```r sigma_vec ``` ``` ## [1] 2 4 6 8 10 ``` ```r n ``` ``` ## [1] 5 1 10 8 2 ``` ] .pull-right[ ```r map2(n, mu_vec, rnorm, sd = sigma_vec) ``` ``` ## [[1]] ## [1] 3.747106 -4.731081 6.500376 -22.013180 ## [5] -7.273418 ## ## [[2]] ## [1] 6.887339 ## ## [[3]] ## [1] 8.379113 8.168874 4.680082 11.516717 ## [5] 14.566685 8.270954 3.709349 9.603832 ## [9] 16.051026 6.281314 ## ## [[4]] ## [1] 11.914212 7.836439 8.116733 7.376050 ## [5] 15.138203 11.171446 13.894821 8.822398 ## ## [[5]] ## [1] 16.07507 26.35510 ``` ] --- # map2() - Example 2 * Apply to nested data! .pull-left[ ```r library(magrittr) nested_df %<>% mutate(model = map(data, ~lm(bill_length_mm~bill_depth_mm, data = .))) nested_df ``` ``` ## # A tibble: 3 x 3 ## species data model ## <fct> <list> <list> ## 1 Adelie <tibble [152 x 7]> <lm> ## 2 Gentoo <tibble [124 x 7]> <lm> ## 3 Chinstrap <tibble [68 x 7]> <lm> ``` ] .pull-right[ ```r nested_df %>% mutate(prediction = map2(model, data, predict)) ``` ``` ## # A tibble: 3 x 4 ## species data model prediction ## <fct> <list> <list> <list> ## 1 Adelie <tibble [152 x 7]> <lm> <dbl [152]> ## 2 Gentoo <tibble [124 x 7]> <lm> <dbl [124]> ## 3 Chinstrap <tibble [68 x 7]> <lm> <dbl [68]> ``` ] --- # Let's `walk()` together * What if we don't want to have data? * Just want to do something like plotting or write files. ```r plot_scatter <- function(df){ ggplot(df, aes(x = bill_length_mm, y = bill_depth_mm)) + geom_point() } penguins %>% split(.$species) %>% map(plot_scatter) -> plots ``` --- # walk() * What's the diffence from ggplot facets? ```r walk(plots, print) ``` <img src="lec3_files/figure-html/unnamed-chunk-37-1.png" width="25%" /><img src="lec3_files/figure-html/unnamed-chunk-37-2.png" width="25%" /><img src="lec3_files/figure-html/unnamed-chunk-37-3.png" width="25%" /> --- # Split the file ```r penguins %>% group_by(species) %>% group_walk(~ write_csv(.x, paste0("data/", .y$species, ".csv"))) ``` --- # Read data .pull-left[ ```r library(fs) files <- dir_ls(path = "data", glob = "*csv") files ``` ``` ## data/Adelie.csv data/Chinstrap.csv ## data/Gentoo.csv ``` ```r adelie <- read_csv("data/Adelie.csv") chinstrap <- read_csv("data/Chinstrap.csv") gentoo <- read_csv("data/Gentoo.csv") all_three <- bind_rows(adelie, chinstrap, gentoo, .id = "species") ``` ] .pull-right[ ```r all_three %>% head() ``` ``` ## # A tibble: 6 x 8 ## species island bill_length_mm bill_depth_mm ## <chr> <chr> <dbl> <dbl> ## 1 1 Torgersen 39.1 18.7 ## 2 1 Torgersen 39.5 17.4 ## 3 1 Torgersen 40.3 18 ## 4 1 Torgersen NA NA ## 5 1 Torgersen 36.7 19.3 ## 6 1 Torgersen 39.3 20.6 ## # ... with 4 more variables: ## # flipper_length_mm <dbl>, body_mass_g <dbl>, ## # sex <chr>, year <dbl> ``` ] --- # Read data with `map_dfr()` ```r map_dfr(files, read_csv, .id = "species") ``` ``` ## # A tibble: 344 x 8 ## species island bill_length_mm bill_depth_mm ## <chr> <chr> <dbl> <dbl> ## 1 data/Adel~ Torger~ 39.1 18.7 ## 2 data/Adel~ Torger~ 39.5 17.4 ## 3 data/Adel~ Torger~ 40.3 18 ## 4 data/Adel~ Torger~ NA NA ## 5 data/Adel~ Torger~ 36.7 19.3 ## 6 data/Adel~ Torger~ 39.3 20.6 ## 7 data/Adel~ Torger~ 38.9 17.8 ## 8 data/Adel~ Torger~ 39.2 19.6 ## 9 data/Adel~ Torger~ 34.1 18.1 ## 10 data/Adel~ Torger~ 42 20.2 ## # ... with 334 more rows, and 4 more variables: ## # flipper_length_mm <dbl>, body_mass_g <dbl>, ## # sex <chr>, year <dbl> ``` --- class: middle, center, inverse # Thanks!